The Sentebiolab Senteligo™️ SARS-CoV-2 (COVID-19) Multiplex qPCR Detection Kit is an in vitro diagnostic test, based on real-time polymerase chain reaction technology. It tests for the presence or absence of ribonucleic acid (RNA) of SARS-CoV-2 coronavirus, specifically, in lower and upper respiratory tract samples from patients suspected of COVID-19 viral infection.
The Sentebiolab Senteligo™️ SARS-CoV-2 (COVID-19) Multiplex qPCR Detection test
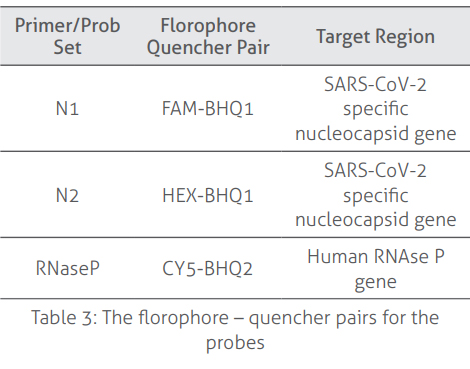
includes a Primer-Probe set inside the Reaction Mix that contains primers and probes
for human RNaseP and viral N1 and N2 regions. The internal control (primer/probe
set for human RNaseP gene) is used to identify possible qPCR inhibition, confirm
the integrity of the reagents, and verify the quality of RNA extraction from patient
material. RNaseP primers and probes included in the kit recognize the human RNaseP
gene. The primers and probes for viral N1 and N2 regions specifically identify two
distinct regions on the nucleocapsid (N) gene of the SARS-CoV-2 virus. These primers and probes do not recognize the SARS-CoV virus, whose genetic structure
is highly similar to SARS-CoV-2
Kit components
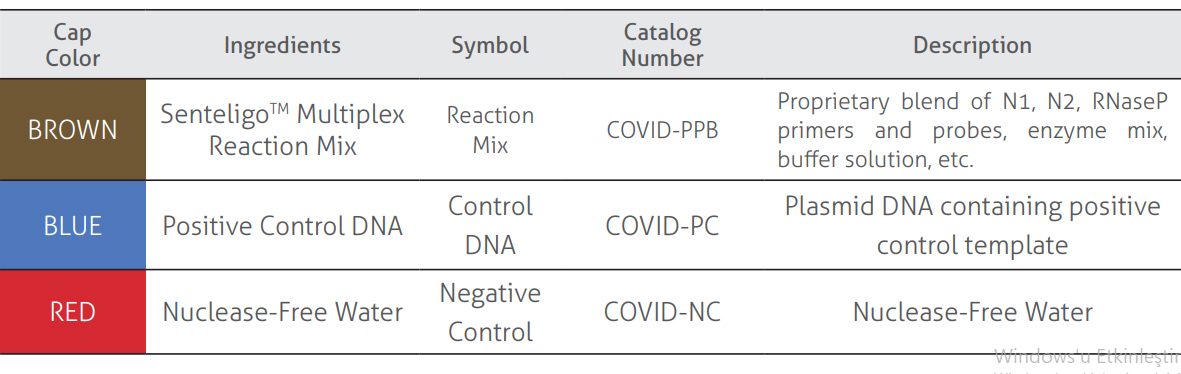
The test has been developed to be used as a single one-step reverse transcription qPCR test for each patient sample, utilizing PCR amplification of the targets (N1, N2 and RNaseP) by FAM, HEX and Cy5 labelled probes. The reaction time is approximately 1 hour, it may differ slightly depending on the qPCR device used. The Kit components test contains internal controls that monitor the performance of the test. The Reaction Mix also contains a propriety mix of Reverse Transcriptase and DNA Polymerase and the necessary buffer solutions and reagents such as nucleic acids in order to perform the qRTPCR reaction. All the reagents are premixed for ease-of-use.
Features & Advantages
- Fast Results
- High-Accuracy (%99)
- Worldwide Acceptance
- Enzyme Include Options
- Minimize Human Error & Contamination
- High Specificity & Sensitivity
- Optimized primer/probe sequences to minimize falsepositives
- Internal control
- to check for RNA quality to minimize false-negatives
- to verify the enzyme and other reagents - 100% inclusivity of published SARS-CoV-2 sequences in unmutated targeted regions
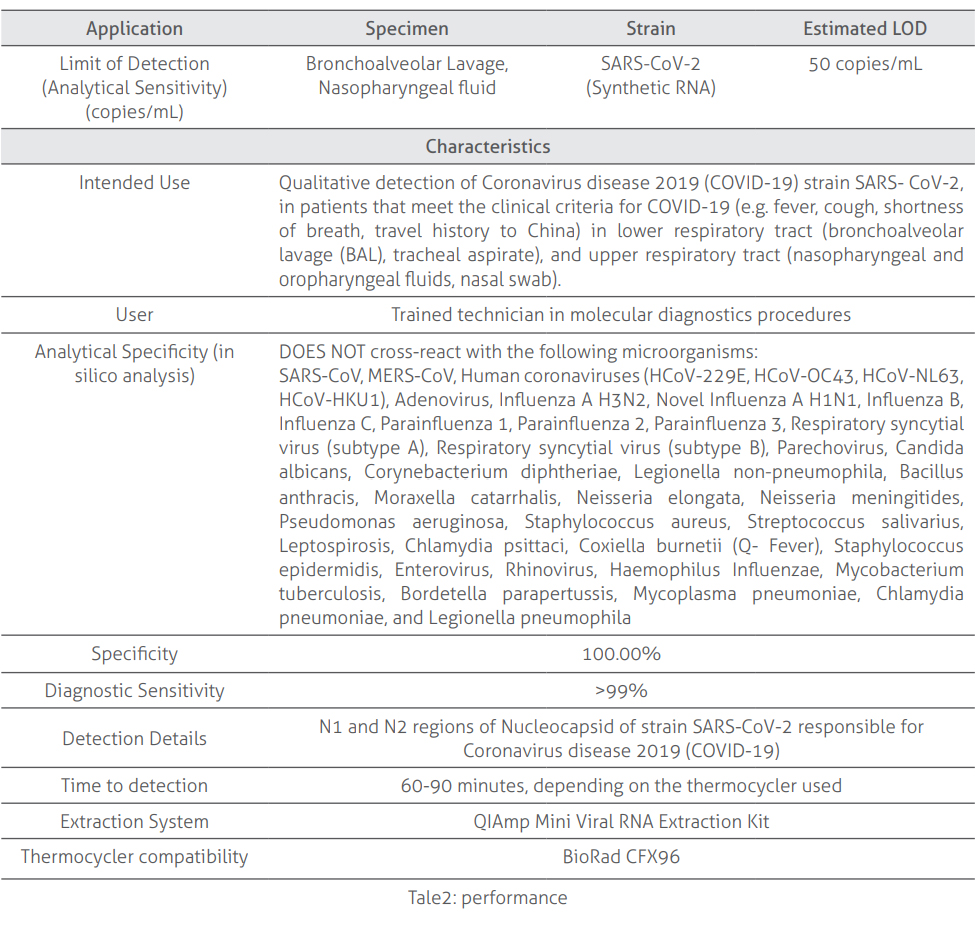
Performance Table
Sorage Conditions
- The kit components should be stored at -20°C.
- Repeated thawing and freezing of components (more than two times) should be avoided, as this might affect the performance of the assay. After thawing, the reagents should be frozen in multiple aliquots if they are to be used intermittently.
- Protect the Reaction Mix from light
since the probes are light-sensitive
and stored in brown vials. Aliquoted Reaction Mixes also need to be
protected from light.
qPCR Protocol
NOTE: All work should be carried out on ice/cold plate. NOTE: All necessary biosecurity measures should be taken when working with patient samples.
1. This kit is a multiplex PCR kit.
The test has been developed to be used as a single one-step reverse transcription qPCR test for each patient sample, utilizing PCR amplification of the targets (N1, N2 and RNaseP) by FAM, HEX and Cy5 labelled probes.2. For each study, a negative control (NC) and a positive control (PC) reaction should be used: a. For negative controls, Negative Control should be added to the wells/tubes instead of the patient sample. The purpose of using a negative control is to determine whether there is nucleic acid contamination in any of the materials used for the amplification. A negative control reaction is mandatory in order to avoid false-positive evaluation in patient samples. No amplification should occur in negative control samples unless there is DNA contamination in any of the test materials. b. For positive controls, Positive Control should be added to the wells/tubes. The purpose of using a positive control is to prevent false-negative cases that may arise from study errors. In order to avoid false-negative evaluations in patient samples, positive control must be studied. Amplification should be observed in positive control samples to ensure that the reaction worked well.
NOTE: The protocol shown below is the optimized protocol for the BioRad CFX96 and HealForce qPCR devices. If you use a different device, a preliminary trial should be performed using only negative control and positive control, and the reaction conditions should be optimized in accordance with the recommendations of the manufacturers of the kit and the device used, when necessary.
3. Design the wells of the plate/ strip tubes to work with 3 sets of primers/probes for each sample, and enter this design to the software of the qPCR device.
4. The total volume of the reaction will be 30 µL. Add 25 µL of Reaction Mix to each tube/well.
5. For negative controls (NC), 5 µL of Negative Control (nuclease-free water) should be added to the wells/tubes instead of the patient sample.
6. Pipet 5 µL of patient sample (elution from nucleic acid extraction) to the appropriate well.
7. For positive controls (PC), 5 µL of Positive Control should be added to the wells/tubes.
8. Cover your tube/plate with optically permeable material (cap/ seal).
9. If you are using strip tubes, spin them down. If you are using a 96 well plate, centrifuge the plate shortly (using plate rotor – for 30 sec at 500 g).
10. Create the reaction protocol in the software of the qPCR device with the following steps: 55 oC - 10 min 95 oC - 30 sec 95 oC - 10 sec - 35 cycle 60 oC - 30 sec - 35 cycle
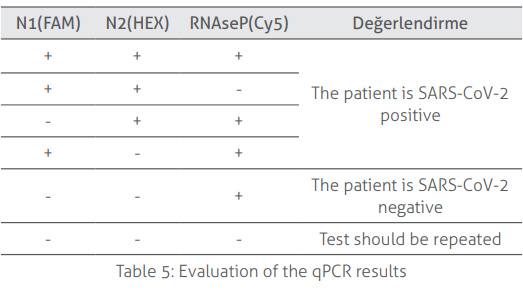
11. Hazırladığınız strip tüp/plate'i qPCR cihazına yerleştirin ve reaksiyonu başlatın.EVALUATION OF THE RESULTS
- For each study, negative results should be obtained from NCs and positive results from PCs. Studies that do not comply with these results must be repeated.
- The reaction curves that are not sigmoidal should be interpreted as negative.
- Positive amplification in the N1 and N2 reactions indicates a positive result despite the lack of concurrent amplification in the RNaseP reaction, since the RNAseP amplification is dependent on the presence of human RNA in the extraction sample, the amount of which is governed by the type of the patient sample and the extraction procedure used. For any other case, positive results should be obtained for RNAseP reactions with an amplification curve or cycle threshold value at or below 35 cycles. Amplification curves greater than 35 cycles for RNAseP are in the uncertainty zone. The presence of a curve, with a Cq at or below 35 cycles, for a sample in the RNAseP, indicates a positive result. Not getting a positive result for RNAseP reaction of a patient sample means that there might be a problem in the RNA isolation stage. Thus, in cases where RNAseP is negative while either N1 or N2 or both are negative, RNA isolation from those samples should be repeated.
- For N1 and N2, a positive result means an amplification curve or cycle threshold value at or below 35 cycles. Amplification curves greater than 35 cycles for N1 or N2 are in the uncertainty zone. The presence of a curve, with a Cq at or below 35 cycles, for a sample in the N1 or N2, indicates a positive result.
- As stated in Table 4, samples with positive results from all N1, N2 and RNAseP sets should be evaluated as SARS-CoV-2 positive; while N1 and N2 negative and RNAseP positive results should be evaluated as SARS-CoV-2 negative. Samples with a positive result from only one of N1 or N2 should be retested.a Cq at or below 35 cycles, for a sample in the N1 or N2, indicates a positive result.
- As stated in Table 4, samples
with positive results from all N1,
N2 and RNAseP sets should be
evaluated as SARS-CoV-2 positive;
while N1 and N2 negative and
RNAseP positive results should be
evaluated as SARS-CoV-2 negative.
Samples with a positive result from
only one of N1 or N2 should be
retested.
Evaluation of the qPCR results